
N.ğinally, find Node Color under Unused Properties and double-click it. For example, set minimum to 0.5 and maximum to 60. Iv.Ědjust the minimum and maximum values denoted by red squares (double-click on squares for precision, otherwise slide squares up or down). M.ğind Edge Opacity under Unused Properties and double-click it. For example, set minimum to 0.5 and maximum to 20.
Iii.Ĝlick on the graph next to the Graphical View property to launch the Continuous Editor.
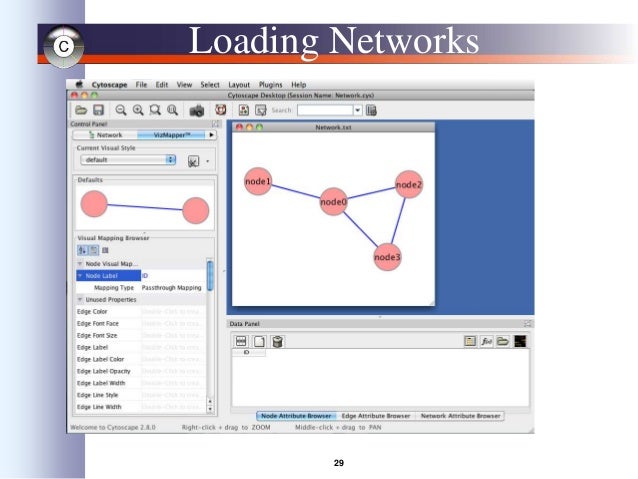
L.ğind Edge Line Width under Unused Properties and double-click it. Drag the leftmost adjustable triangle all the way to the left and likewise drag the rightmost triangle to the right until it stops. Double-click the two rightmost triangles and set their colors to pure blue (0,0,255). Double-click the two leftmost triangles and set their colors to pure red (255,0,0). There are two fixed triangles, one on each end, and two adjustable triangles. Iii.Ĝlick on the black-to-white gradient next to Graphical View to launch a Gradient Editor. Select Continuous Mapper for Mapping Type. Under Unused Properties find Edge Color and double-click it. Minimize the Node Label property by selecting the minus icon. All nodes should now be relabeled according to the original data labels. Next to Node Label, click ID and select Column 3. Go back to the Node tab and change the NODE_BORDER_COLOR property to black and increase the NODE_LINE_WIDTH to 2. Iii.Ĝhange the background color to white. Select the Global tab in the bottom right. Click in the Defaults window (shows a source and target pair with a blue background). In the Control Panel, select the VizMapperTM tab. H.Ĝhange global properties of the network: Go to Select File(s) and locate AutoSOME output file (5) containing all nodes and attributes (see Files written to disk in the manual) or use the tutorial dataset. Go to File> Import> Attribute from Table (Text/MS Excel).į. Finally, click on Column 3 in the data Preview window to activate it (it will turn blue).ĭ. Set Source Interaction to Column 1 and Target Interaction to Column 2. Go to Select File(s) and locate AutoSOME output file (4) containing all edges (see Files written to disk in the manual) or use the tutorial dataset.Ĭ. Go to File> Import> Network from Table (Text/MS Excel).ī.
#CYTOSCAPE 3 TUTORIAL SERIES#
cell samples or time series of a microarray dataset)ĥ) Within Cytoscape (All screenshots below taken from Cytoscape 2.6.0):Ī. (Note that only vertical data vectors are clustered (e.g.
#CYTOSCAPE 3 TUTORIAL MANUAL#
*You may skip steps 1-3 and use the tutorial datasets provided below.ġ) Import your input file into AutoSOME, set fields and adjust input (see the manual for instructions).Ģ) Select Fuzzy Cluster Networks and choose distance metric. New! AutoSOME fuzzy clustering can now be performed directly in Cytoscape using the clusterMaker plugin. Below, a protocol is given that requires Cytoscape. To display fuzzy cluster networks, a network visualization tool needs to be installed on your computer. A powerful application of this approach is the visualization of differences between cell lines on the basis of differential gene expression. AutoSOME Web Portal: Fuzzy Cluster Networksįuzzy cluster networks highlight the fuzzy relationships among clustered data points using an intuitive two-dimensional network display.


 0 kommentar(er)
0 kommentar(er)
